Conformer Search (GOAT)¶
For flexible molecules, it is important to identify the low energy conformational space to provide
the most accurate model of the investigated system. The global optimizer algorithm (GOAT) within ORCA
can help to automatically find the global minimum structure and the ensemble around it. GOAT can be
envoked by the simple input keyword GOAT:
!GOAT XTB
*XYZFILE 0 1 inp.xyz
Note
As a GOAT run involves a large number of geometry optimizations, it is recommended to parallelize
the calculation, e.g. via the PAL4 keyword. Further, specifically for large systems, the usage of
fast semi-empirical methods like GFN2-xTB speeds up the calculation. Nevertheless, GOAT can be used
with any method in ORCA that is capable of optimizing geometries.
Example: Diclofenac¶
As an example, we will use to find the global minimum conformation of Diclofenac starting from the PubChem structure.
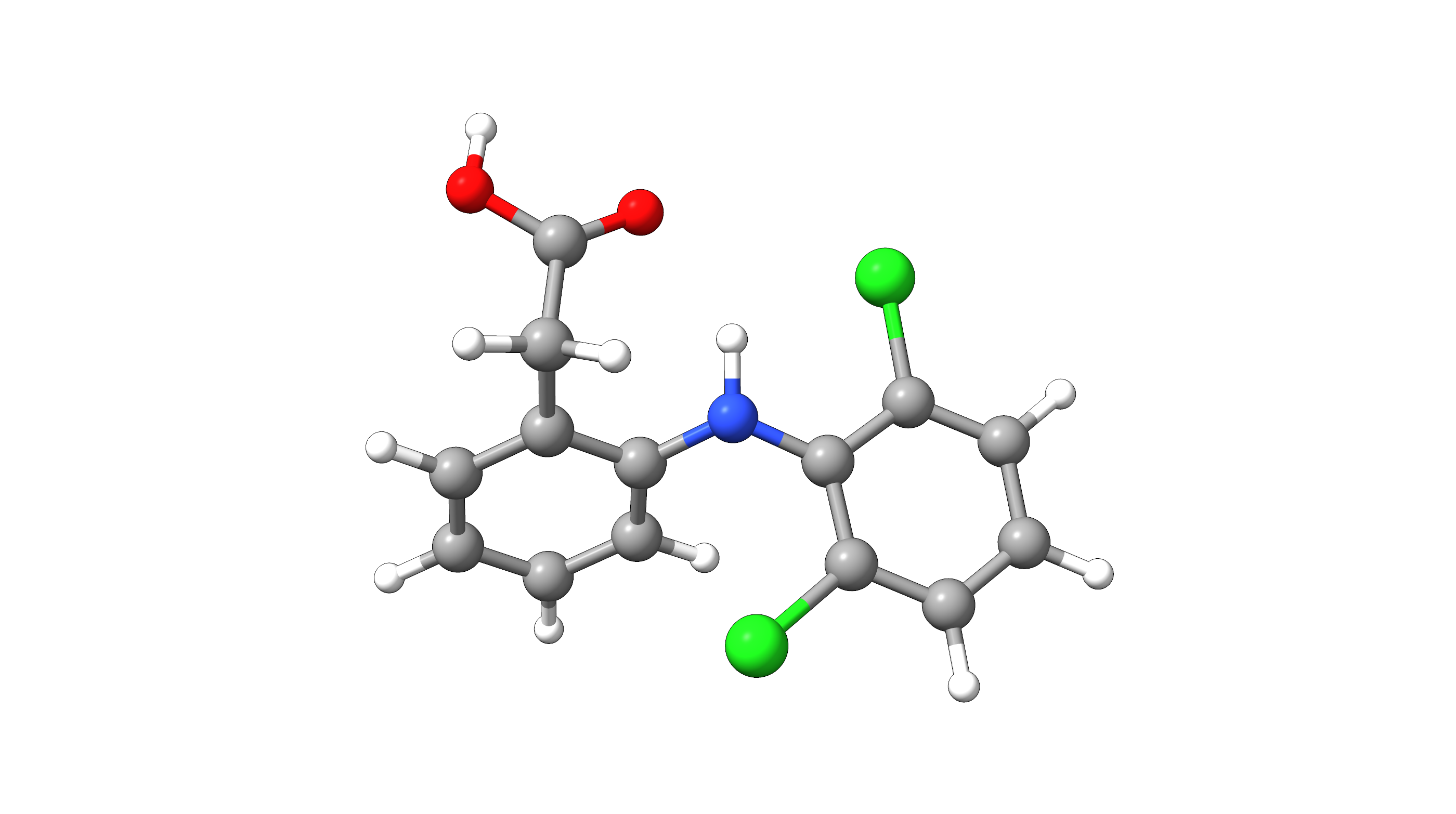
Figure: Diclofenac¶
The input looks like:
!GOAT XTB
*XYZFILE 0 1 diclofenac.xyz
After the successful GOAT run, the global minimum structure is stored in the
basename.globalminimum.xyz file and the full structure ensemble in the basename.finalensemble.xyz
file. In this case, GOAT found 17 unique conformers. The corresponding energies and weights are
reported in the output file:
------------------------------
_
| |
__ _ ___ __ _| |_
/ _` |/ _ \ / _` | __|
| (_| | (_) | (_| | |_
\__, |\___/ \__,_|\__|
__/ |
|___/
A Global Optimizer Algorithm
------------------------------
[...]
Global minimum found!
Writing structure to diclofenac.globalminimum.xyz
# Final ensemble info #
Conformer Energy Degen. % total % cumul.
(kcal/mol)
------------------------------------------------------
0 0.000 1 75.54 75.54
1 0.976 1 14.56 90.10
2 1.991 1 2.62 92.72
3 2.028 1 2.46 95.18
4 2.413 1 1.29 96.47
5 2.572 1 0.98 97.45
6 2.621 1 0.91 98.36
7 2.799 1 0.67 99.03
8 3.339 1 0.27 99.30
9 3.420 1 0.24 99.53
10 3.781 1 0.13 99.66
11 3.827 1 0.12 99.78
12 4.027 1 0.08 99.86
13 4.373 1 0.05 99.91
14 4.614 1 0.03 99.94
15 4.624 1 0.03 99.97
16 4.752 1 0.02 100.00
17 5.951 1 0.00 100.00
Conformers below 3 kcal/mol: 8
Lowest energy conformer : -55.771201 Eh
Sconf at 298.15 K : 1.83 cal/(molK)
Gconf at 298.15 K : -0.17 kcal/mol
Writing final ensemble to diclofenac.finalensemble.xyz
We see, that the conformer space of Diclofenac is dominated by the two conformers 1 and 2, with their weights summing up to 90.1 %. The 10 lowest conformers look like:
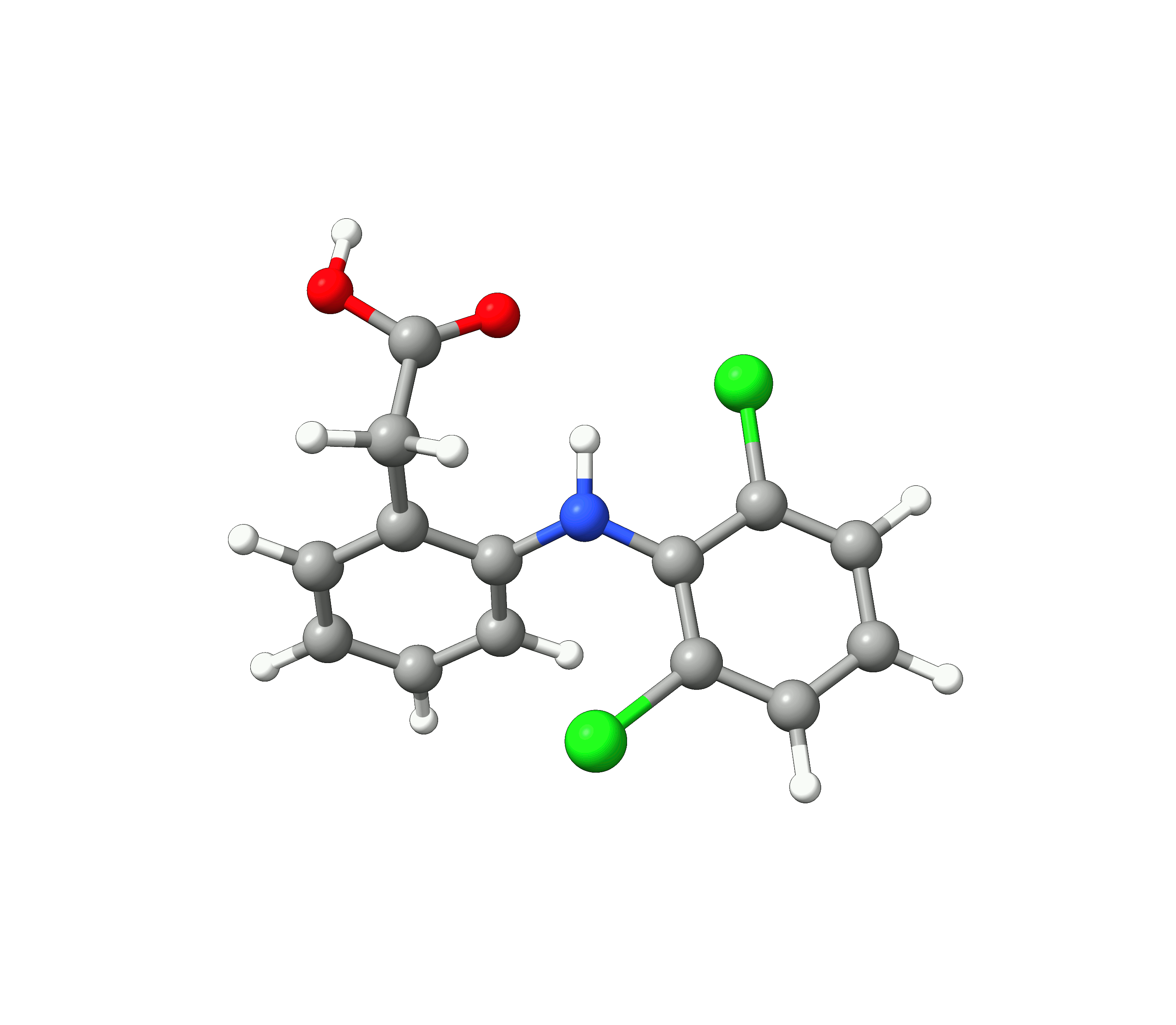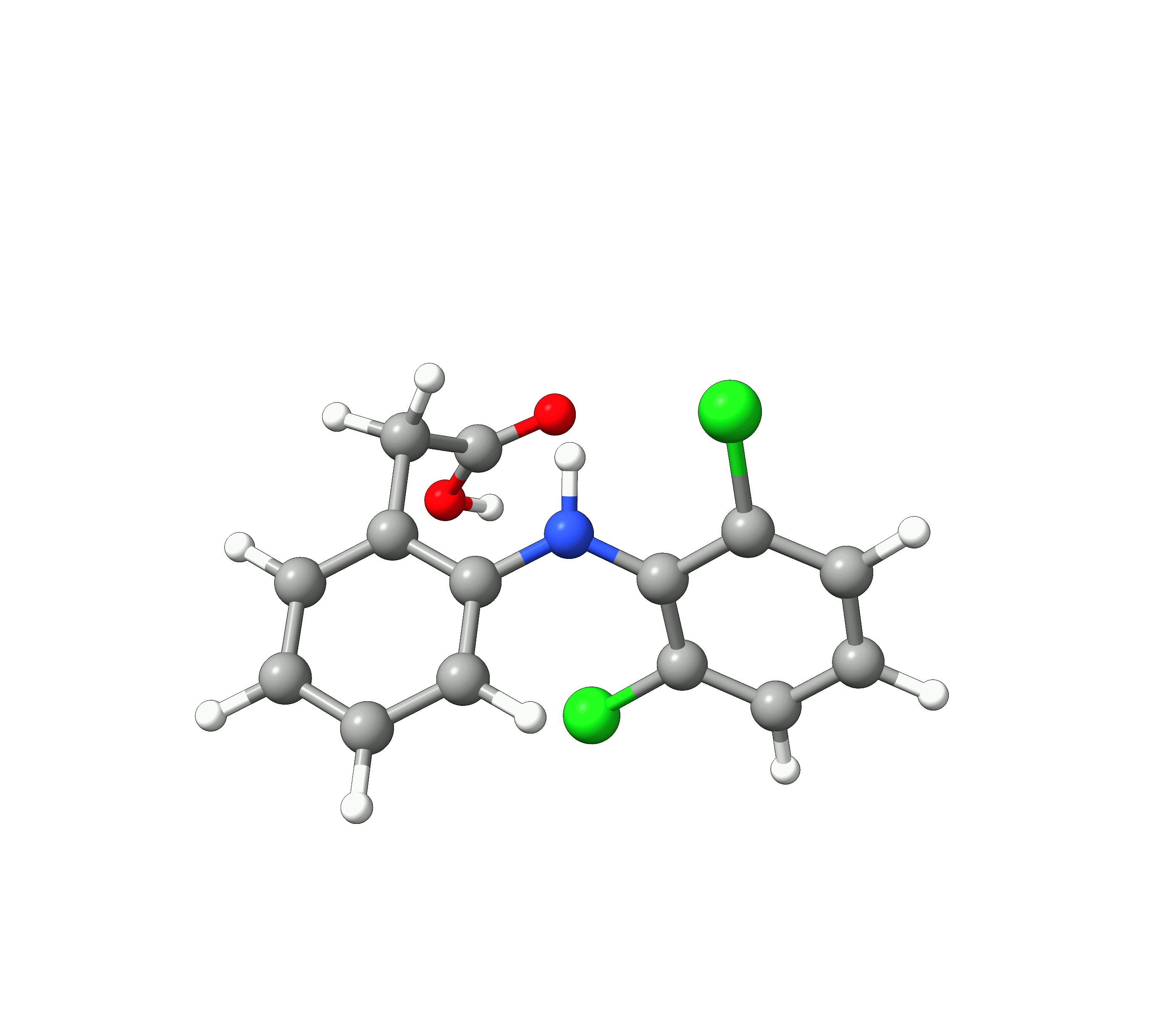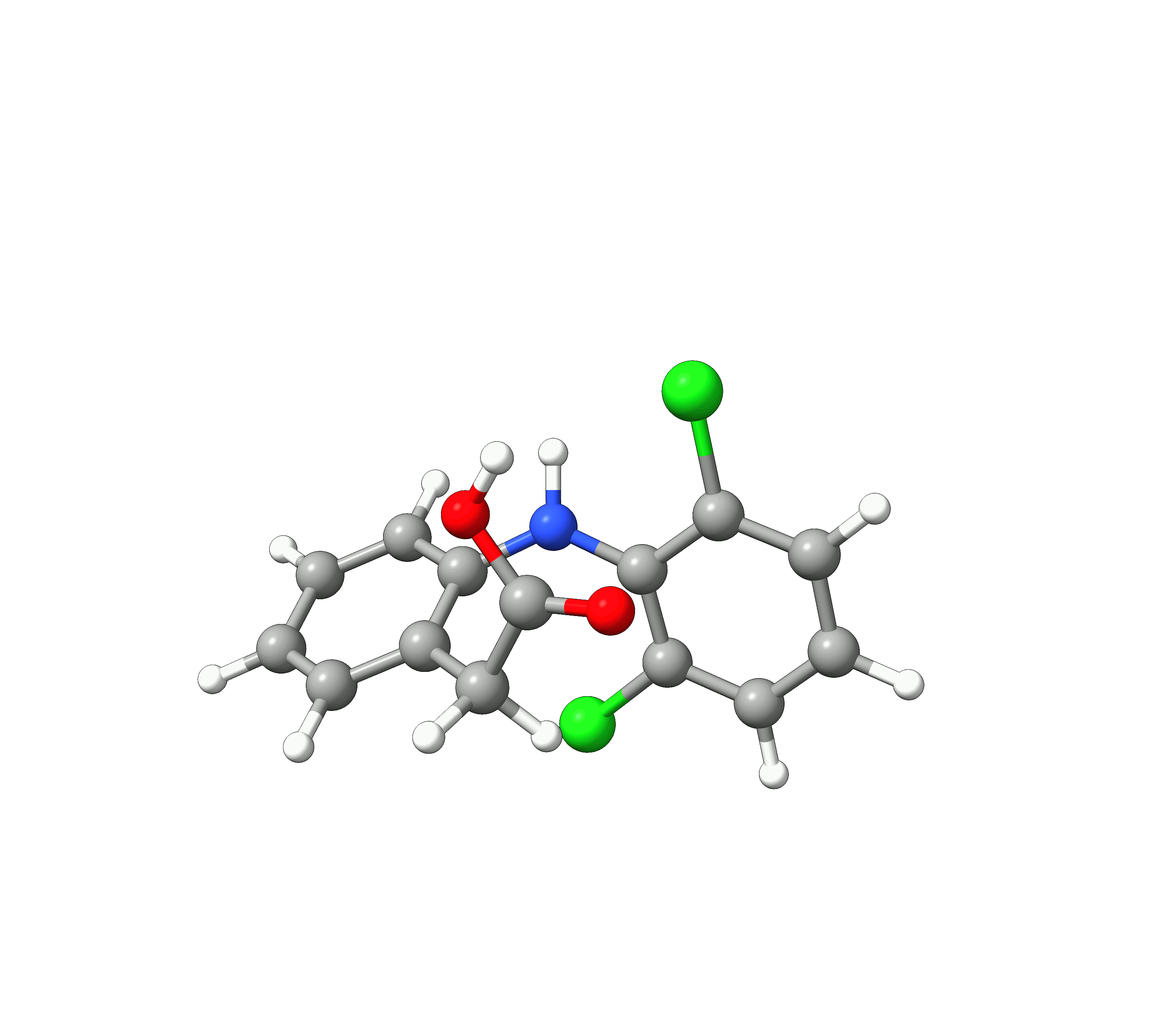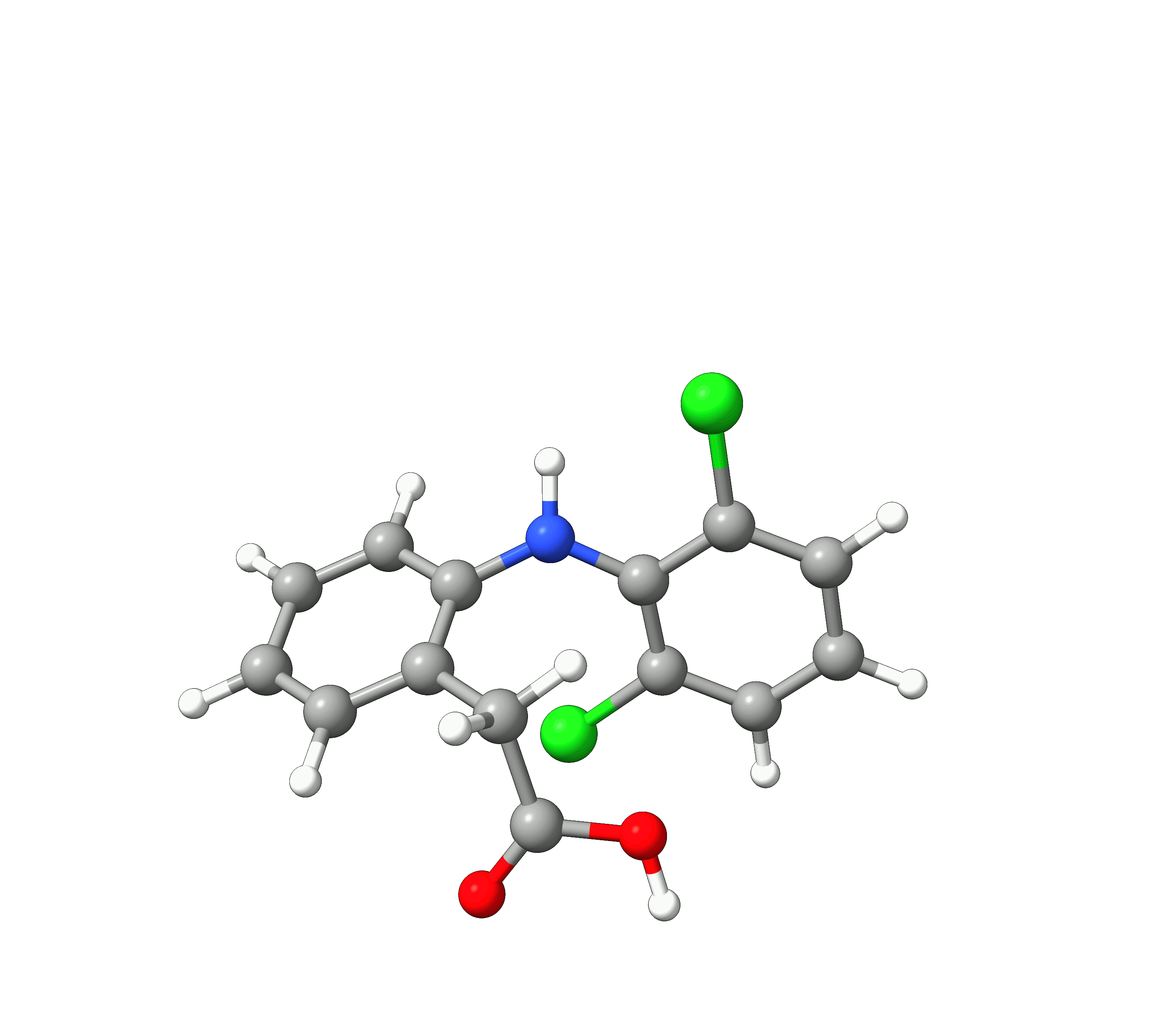
Figure: The 10 lowest conformers of Diclofenac found by GOAT.¶
Structures¶
Diclofenac from PubChem 3033
30
Cl 1.95410 1.15000 -2.50780
Cl 1.13770 -1.63920 2.11360
O -3.26200 -2.92840 -1.06470
O -2.79060 -1.91080 0.90920
N 0.26790 -0.20510 -0.39900
C -2.06400 0.51390 -0.37690
C -0.73130 0.71780 -0.01920
C -2.47610 -0.68300 -1.17030
C 1.65710 -0.24820 -0.17950
C -3.03820 1.43500 0.00810
C -0.37280 1.84290 0.72340
C -2.67970 2.56000 0.75060
C -1.34700 2.76400 1.10830
C 2.53530 0.34770 -1.09180
C 2.17400 -0.88650 0.95340
C -2.84800 -1.87490 -0.31230
C 3.91240 0.30580 -0.87390
C 3.55110 -0.92850 1.17130
C 4.42030 -0.33240 0.25760
H -1.70860 -0.97920 -1.89300
H -3.36140 -0.42660 -1.76760
H -0.08610 -1.11460 -0.67800
H -4.08120 1.28850 -0.26040
H 0.65690 2.02780 1.01670
H -3.43820 3.27690 1.05110
H -1.06830 3.63990 1.68680
H 4.60370 0.76540 -1.57580
H 3.96350 -1.42150 2.04800
H 5.49250 -0.36510 0.42740
H -3.50250 -3.70110 -0.51020